Avogadro APBS
Jump to navigation
Jump to search
Avogadro2 supports APBS through the apbs extension.
Workflow
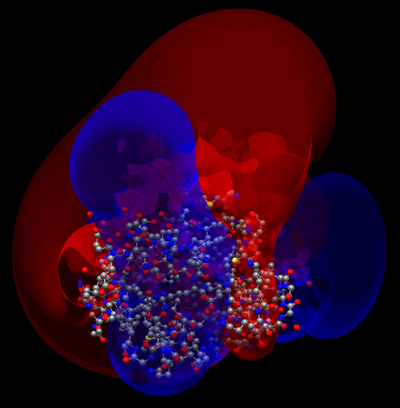
The following steps show how to calculate an electrostatic potential surface using APBS through Avogadro. First download the PDB file for the protein of interest. In this example we use 1MYK.pdb (available from the AvogadroData here).
- Create PQR file for the input molecule.
- This requires that the `pdb2pqr` script be installed on the system.
- Under the Extensions->APBS menu click on "Run PDB2PQR".
- Select the input PDB file (e.g. 1MYK.pdb) and the AMBER force field.
- Click 'Run' and select an output file name (e.g. 1MYK.pqr).
- Create apbs.in file.
- Under the Quantum->Input Generators menu click on "APBS".
- Enter the input file name (e.g. 1MYK.pqr) and then click "Write files to disk...".
- Run apbs.
- On the command line run "apbs apbs.in" in the same directory that you saved the input file (e.g. apbs.in) and molecule file (e.g. 1MYK.pqr).
- Open the output file.
- Under the Extensions->APBS menu click on "Open Output File".
- Navigate the potential file from apbs (e.g. "pot-PE0.dx") and click open. The positive and negative potential surfaces will be created.
- Enable the "Meshes" rendering engine to display the meshes in the render view.