GSoC Ideas 2017
Guidelines
Open Chemistry is an umbrella for projects in chemistry, materials science, biochemistry, and related areas. We intend to concentrate mainly on projects to improve Avogadro 2, cclib, 3DMol, and Open Babel. We have gathered a pool of interested mentors together who are seasoned developers in each of these projects. We welcome original ideas in addition to what's listed here - please suggest something interesting for open source chemistry!
Adding Ideas
When adding a new idea to this page, please try to include the following information:
- A brief explanation of the idea.
- Expected results/feature additions.
- Any prerequisites for working on the project.
- Links to any further information, discussions, bug reports etc.
- Any special mailing lists if not the standard mailing list for the project
- Your name and email address for contact (if willing to mentor, or nominated mentor).
Proposal Guidelines
Students need to write and submit a proposal, we have added the applying to GSoC page to help guide our students on what we would like to see in those proposals.
Avogadro 2 Project Ideas
Avogadro 2 is a chemical editor and visualization application, it is also a set of reusable software libraries written in C++ using principles of modularity for maximum reuse. We offer permissively licensed, open source, cross platform software components in the Avogadro 2 libraries, along with an end-user application with full source code, and binaries.
Project: Integrate with VTK: Volume Rendering and Charts
Brief explanation: Volume rendering is great for visualizing electronic structure, and VTK also brings charts for quantitative data
Expected results: The Visualization Toolkit (VTK) features a number of rendering techniques for 3D geometry (including volume rendering), and 2D charts (line, points, parallel coordinates, scatter plot matrices). These are all written using a recently updated OpenGL backend, and recent improvements to the Qt5 code in VTK make integration with Avogadro much simpler. This project would focus primarily on exposing the QVTKOpenGLWidget to Avogadro 2, and then exposing volume rendering for 3D electronic structure data, and tables in the charts. Integration would involve developing some interface classes that make it easy to interact with the VTK classes from Avogadro code, and some concrete implementations showing data in these widgets.
Prerequisites: Experience in C++, some experience with OpenGL ideal, but not necessary.
Mentor: Marcus D. Hanwell (marcus dot hanwell at kitware dot com).
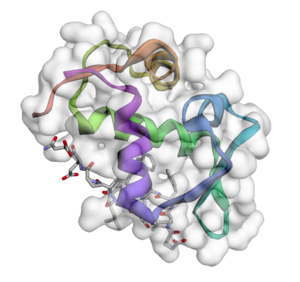
Project: Biological Data Visualization
Brief explanation: Support for biological data, representations, and visualization
Expected results: Add support for molecular fragments on top of the molecule model, extending this to residues, and supporting reading/writing this secondary structure (e.g., PDB format). Additional rendering modes for secondary biological structures (i.e. ribbons, cartoons, etc.), building up a biomolecule from residues, and adding residue labels. Code and algorithms may be adapted from 3DMol.js. Since biological molecules are often large (10^3 to 10^6 atoms and bonds), such implementations should be highly efficient and optimized, adopting symmetry and other techniques to improve interactivity and rendering performance.
Prerequisites: Experience in C++, some experience with OpenGL and an biochemistry ideally, but not necessary.
Mentor: Marcus D. Hanwell (marcus dot hanwell at kitware dot com) or Geoffrey Hutchison (geoffh at pitt.edu)
Project: Molecular Dynamics
Brief explanation: Improve support for running, reading, and analyzing molecular dynamics simulations in Avogadro 2
Expected results: Code for running MD simulations in common packages (e.g., OpenMM, GROMACS, etc.) including extracting parameters and atom types from Open Babel. Initial support for reading in basic trajectories from XYZ files, and static .gro files for GROMACS already exists. Extend this to more fully support the needs of molecular dynamics, reading in trajectory files, ideally loading in time steps on demand for large files rather than loading the entire file in up front (as is currently the case). Investigate whether compression techniques (e.g., delta compression) can improve reading and rendering performance. Investigate ways to support generating input, and dealing with large systems (over one million particles). Add support for characterizing particle movement (e.g., pair-wise distribution functions), rare events, and visualizing these in addition to simple trajectory animations.
Prerequisites: Experience in C++, some experience with OpenGL and an MD code ideal, but not necessary.
Mentor: Marcus D. Hanwell (marcus dot hanwell at kitware dot com).
Project: Scripting Bindings
Brief explanation: Implement an embedded scripting language (e.g., Python or JavaScript) in Avogadro 2
Expected results: Create bindings for the C++ libraries in Python or JavaScript / QtScript. This should allow an embedded scripting console as well as support for implementing modular extensions (tools, rendering, etc.) in Python or JavaScript. A Boost.Python implementation existed in Avogadro v1, and some rudimentary support has been re-implemented with the new code base. An ideal solution would connect to QML and Qt to allow scripting to add menu items, windows, etc. and provide documentation and example scripts. The interface should be maintainable as new classes and methods are added.
Prerequisites: Experience in C++ and Python or JavaScript, some experience with SWIG, Boost.Python, or similar packages (pybind11, SIP, PySide, etc.) suggested.
Mentor: Geoff Hutchison (geoffh at pitt dot edu)
Project: Integrate with RDKit
Brief explanation: Integrate the RDKit toolkit into Avogadro for conformer sampling and force field optimization
Expected results: RDKit is a BSD-licensed cheminformatics toolkit with a wide range of features useful for Avogadro 2. Most notably, RDKit offers efficient and accurate 3D coordinate generation, conformer sampling, and force field optimization. Implement a connection between Avogadro objects (molecules and atoms) and RDKit objects and implement conformer sampling and force field optimization code.
'Prerequisites: Experience in C++, some experience with Python will be helpful.
Mentor: Geoff Hutchison (geoffh at pitt dot edu)
cclib Project Ideas
cclib is an open source library, written in Python, for parsing and interpreting the results of computational chemistry packages. The goals of cclib are centered around the reuse of data obtained from these programs and contained in output files.
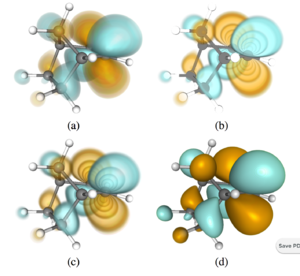
Project: Advanced Analysis of Quantum Chemistry Data
Brief explanation: Implement additional analysis and quantum calculation methods, including ELF (electron localization function), AIM (Bader's Atoms-in-Molecules) techniques, and/or DDEC6 atomic charges.
Expected results: The current cclib library offers some calculation methods, including fragment analysis and some charge models. Many modern analysis techniques exist to partition electron density, including computing gradients, Laplacians, ELF (electron localization function), Bader's AIM analysis, etc. Similarly multiple partial charge assignment methods exist and can be implemented, including DDEC6.
Suggested Readings:
Prerequisites: Experience with Python and linear algebra (including numpy, scipy), some experience with numerical methods suggested.
Mentor: Geoff Hutchison (geoffh at pitt dot edu)
Project: Refactor parsers
Brief explanation: The main extract() function of each parser are quite long and should be split into smaller functions for maintainability.
Expected results: Ensure test coverage of cclib prior to refactoring, propose a few different approaches and discuss with cclib team, and implement best proposal. Ideally new functions are consistent across parsers and associated docstrings can be used for keeping documentation up-to-date.
Prerequisites: Experience with the Python.
Mentor: Adam Tenderholt (atenderholt at gmail dot com) and possibly Karol Langner (karol.langner at gmail dot com).
Project: Implement new parsers
Brief explanation: There are outstanding issues on Github for new parsers.
Expected results: Generate test data and unit tests, and implement new parsers. For example, allow reading and writing from Molden and AIMALL wfn/wfx files.
Prerequisites: Experience with the Python, and ideally familiarity with computational chemistry programs.
Mentor: Adam Tenderholt (atenderholt at gmail dot com) and possibly Karol Langner (karol.langner at gmail dot com).
Open Babel Project Ideas
Open Babel is an open toolbox for chemistry, designed to speak the many languages of chemical data. It's an open, collaborative project allowing anyone to search, convert, analyze, or store data from molecular modeling, chemistry, solid-state materials, biochemistry, or related areas.
Project: Implement MMTF format
Brief explanation: Implementation of MMTF file format in OpenBabel.
Expected results:' Macromolecular Transmission Format (MMTF) is a new compact binary format to transmit and store biomolecular structural data quickly and accurately (http://mmtf.rcsb.org). Your task is to implement support for this format in the OpenBabel open-source cheminformatics toolkit (http://openbabel.org). Code will be written in C++.
Mentor: Geoff Hutchison (geoffh at pitt dot edu) or David Koes (dkoes at pitt dot edu)
Project: Fast, Efficient Fragment-Based Coordinate Generation
Brief explanation: A critical challenge is generating 3D coordinates for a known molecule. Implement a fragment-based generator to supplement the rule-based algorithm.
Expected results: Many representations in chemistry either store no coordinates (SMILES, InChI, FASTA sequence) or 2D coordinates. Performing calculations requires the full 3D reconstruction of a molecule, ideally with a low-energy conformation. Currently, Open Babel uses a combination of a rule-based approach (i.e., expected geometries) to generate atom-by-atom the 3D coordinates of molecules. Fragments are only used for some ring-based structures. For inorganic and organometallic molecules, the rules may fail.
Importantly, the approach is highly efficient, since fragments can set many atoms at once. The project should generate a library reflecting a balance between efficiency (i.e., many common fragments) and size, as well as an efficient, parallel algorithm for connecting fragments. A knowledge-based fragment approach can also supplement and minimize the need for conformer sampling.
A recent research paper describing one such approach can be found here
Prerequisites: Experience in C++ and linear algebra. Knowledge of statistics (e.g., Bayesian inference, data mining), OpenMP or OpenCL ideal.
Mentor: Geoff Hutchison (geoffh at pitt dot edu) or David Koes (dkoes at pitt dot edu)
3Dmol.js Project Ideas
3Dmol.js is a modern, object-oriented JavaScript library for visualizing molecular data that is forked from GLmol. A particular emphasis is placed on performance.
Project: Implement volumetric rendering in 3Dmol.js
Brief explanation: Volumetric rendering provides a way to visualize volumetric data in more detail than simple isosurfaces.
Expected results: A number of different volumetric rendering techniques will be implemented and evaluated for a variety of molecular data types.
Prerequisites: Familiarity with JavaScript, WebGL and/or OpenGL, and basic matrix algebra.
Mentor: David Koes (dkoes@pitt.edu)
Project: Molecular Dynamics Visualization
Brief explanation: Implement high-performance in-browser visualization of molecular dynamics simulations.
Expected results: Initial support is already present, with support for reading in basic trajectories from XYZ files, and static .gro files for GROMACS. Extend this to more fully support the needs of molecular dynamics, reading in trajectory files, ideally loading in time steps on demand for large files rather than loading the entire file in up front. Investigate whether compression techniques (e.g., delta compression) can improve reading and rendering performance. Investigate ways to support generating input, and dealing with extremely large systems (over one million particles). Add support for characterizing particle movement (e.g., pair-wise distribution functions), rare events, and visualizing these in addition to simple trajectory animations.
Prerequisites: Experience with JavaScript and client-server programming, some experience with OpenGL/WebGL and an MD code ideally, but not necessary.
Mentor: David Koes l (dkoes@pitt.edu).
Miscellaneous Project Ideas
These ideas would likely benefit two or more projects.
Project: GPU and Multi-Core Enabled High Performance Force Field Calculations
Brief explanation: Add integrated molecular mechanics force field simulations in Avogadro 2
Expected results: Currently, Avogadro 2 relies on command-line calls to Open Babel to optimize geometries or perform conformer searching. The Open Babel code supports multiple force fields, but has poor performance. A modern implementation of a force field library would be welcome, including OpenMP and/or OpenCL support for highly parallel calculations. The architecture should support constrained geometry optimizations and multiple optimization techniques (i.e., steepest descent, conjugate gradients, quasi-Newton like L-BFGS) and be modular enough to allow new force field implementations as plugins.
Ideally the code would be implemented in a new library so it can be used by Avogadro, Open Babel, and other codes
Prerequisites: Experience in C++, some experience with OpenMP or OpenCL ideally.
Mentor: Geoff Hutchison (geoffh at pitt dot edu).
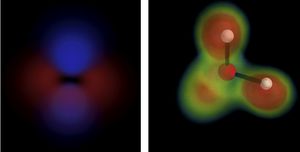
Project: GPU Accelerated Calculation of Molecular Surfaces and QM Data
Brief explanation: Leverage generic GPU/CPU language (e.g., OpenCL or CUDA) to efficiently generate molecular surface data, including as molecular orbitals or electron (spin) density. This may include methods to approximate the molecular surface using compressed forms (e.g., Gaussian spheres). Current code uses multiple CPU cores, but competing codes can produce near-instantaneous rendering using OpenCL: http://www.kieber-emmons.com/Lumo/) Similar code exists in VMD.
Additional performance improvements may come through efficient surface generation techniques used in other work (e.g., using the Euclidian Distance Transform).
Expected results: Generate appropriate kernels that can be used in any language that supports OpenCL (C, C++, Python, etc.) across multiple platforms.
Prerequisites: General programming experience, and ideally experience in chemistry and matrix manipulations.
Suggested Readings:
- http://www.ks.uiuc.edu/Publications/Papers/paper.cgi?tbcode=STON2009
- http://zhanglab.ccmb.med.umich.edu/EDTSurf/
- https://www.cgl.ethz.ch/Downloads/Publications/Papers/2009/Jan09/Jan09.pdf
Mentor: Geoffrey Hutchison (geoffh at pitt dot edu)
Project: OneMol: Google Docs & YouTube for Molecules
Brief explanation: There is a huge need in the research community for improved collaboration tools on web and desktop. OneMol will provide an open API for collaborating on molecular data that both Avogadro and 3Dmol.js will support as reference implementations. OneMol compliant applications will be able to manipulate and view molecular data in real time so that changes made by one client will be propagated to other clients.
File-sharing is a means for sharing data, but it does not share real-time interactions; each user’s data exists in its own isolated environment. Screen-sharing provides a common viewpoint for all participants, but allowing others to interact with the data requires granting access to the host workstation. This approach is needlessly inefficient for the task of collaborating on molecular data, and this inefficiency introduces scalability issues. For example, a simple rotation necessitates a full screen update when the fundamental change in state was a simple change in viewing angles.
The OneMol framework consists of three main components: a client module, embedded in a molecular viewer; a facilitator module that enforces a consistent viewer state between all the clients; and a storage module that stores the raw molecular data. All three modules may coexist on the same machine within the same application. However, we anticipate a more common modality will be to use a publicly hosted facilitator server, since this simplifies network connectivity in the face of firewalls and network address translation.
Expected results: Prototype web services to allow web and/or desktop collaboration using 3DMol as a viewer, likely integrating with existing storage systems (e.g., MongoChem or PQR).
Prerequisites: Experience with scripting, and web services. Interest and experience with databases like MongoDB or DSpace very helpful.
Mentor: David Koes (dkoes@pitt.edu) or Geoffrey Hutchison (geoffh at pitt.edu)